The main aim of collaboration between the National Competence Centre for HPC (NCC HPC) and the Institute of Polymers of SAV (IP SAV) was design and implementation of a pilot software solution for automatic processing of polymer microcapsules images using artificial intelligence (AI) and machine learning (ML) approach. The microcapsules consist of semi-permeable polymeric membrane which was developed at the IP SAV.

Automatic image processing has several benefits for IP SAV. It will save time since manual measurement of microcapsule structural parameters is time-consuming due to a huge number of images produced during the process. In addition, the automatic image processing will minimize the errors which are inevitably connected with manual measurements. The images from optical microscope obtained with 4.0 zoom usually contain one or more microcapsules, and they represent an input for AI/ML process. On the other hand, the images from optical microscope obtained with 2.5 zoom usually contain (three to seven) microcapsules. Herein, a detection of the particular microcapsule is essential.
The images from optical microscope are processed in two steps. The first one is a localization and detection of the microcapsule, the second one consists of a series of operations leading to obtaining structural parameters of the microcapsules.
Microcapsule detection
YOLOv5 model with pre-trained weights from COCO128 dataset was employed for microcapsule detection. Training set consisted of 96 images, which were manually annotated using graphical image annotation tool LabelImg [3]. Training unit consisted of 300 epochs, images were subdivided into 6 batches per 16 images and the image size was set to 640 pixels. Computational time of one training unit on the NVIDIA GeForce GTX 1650 GPU was approximately 3.5 hours.
The detection using the trained YOLOv5 model is presented in Figure 1. The reliability of the trained model, verified on 12 images, was 96%, with the throughput on the same graphics card being approximately 40 frames per second.
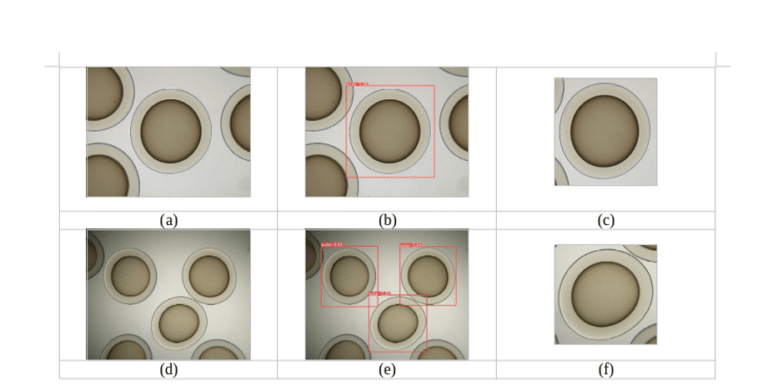
Figure 1: (a) microcapsule image from optical microscope (b) detected microcapsule (c) cropped detected microcapsule for 4.0 zoom, (d) microcapsule image from optical microscope (e) detected microcapsule (f) cropped detected microcapsule for 2.5 zoom.
Measurement of microcapsule structural parameters using AI/ML
The binary masks of inner and outer membrane of the microcapsules are created individually, as an output from the deep-learning neural network of the U-Net architecture [4]. This neural network was developed for image processing in biomedicine applications. The first training set for the U-Net neural network consisted of 140 images obtained from 4.0 zoom with the corresponding masks and the second set consisted of 140 images obtained from 2.5 zoom with the corresponding masks. The training unit consisted of 200 epochs, images were subdivided into 7 batches per 20 images and the image size was set to 1280 pixels (4.0 zoom) or 640 pixels (2.5 zoom). The 10% of the images were used for validation. Reliability of the trained model, verified on 20 images, exceeded 96%. Training process lasted less than 2 hours on the HPC system with IBM Power 7 type nodes, and it had to be repeated several times. Obtained binary masks were subsequently post-processed using fill-holes [5] and watershed [6] operations, to get rid of the unwanted residues. Subsequently, the binary masks were fitted with an ellipse using scikit-image measure library [7]. First and second principal axis of the fitted ellipse are used for the calculation of the microcapsule structural parameters. An example of inner and outer binary masks, and the fitted ellipses is shown in Figure 2.
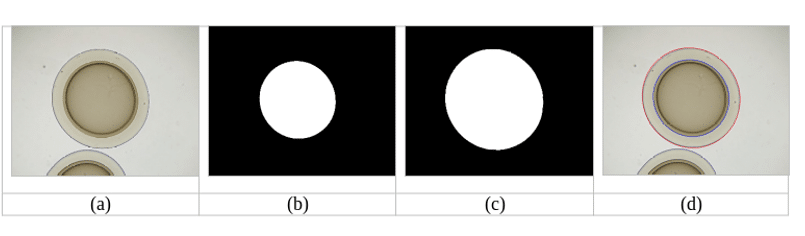
Figure 2: (a) input image from optical microscope (b) inner binary mask (c) outer binary mask (d) output image with fitted ellipses.
Structural parameters obtained by our AI/ML approach (denoted as “U-Net“) were compared to the ones obtained by manual measurements performed at the IP SAV. A different model (denoted as “Retinex”) was used as another independent source of reference data. The Retinex approach was implemented by RNDR. Andrej Lúčny, PhD. from the Department of Applied Informatics of the Faculty of Mathematics, Physics and Informatics in Bratislava. This approach is not based on the AI/ML, the ellipse fitting is performed by the aggregation of line elements with low curvature using so-called retinex filler [8]. The Retinex approach is a good reference due to its relatively high precision, but it is not fully automatic, especially for the inner membrane of the microcapsule.
Figure 3 summarizes a comparison between the three approaches (U-Net, Retinex, UP SAV) to obtain the 4.0 zoom microcapsule structural parameters.
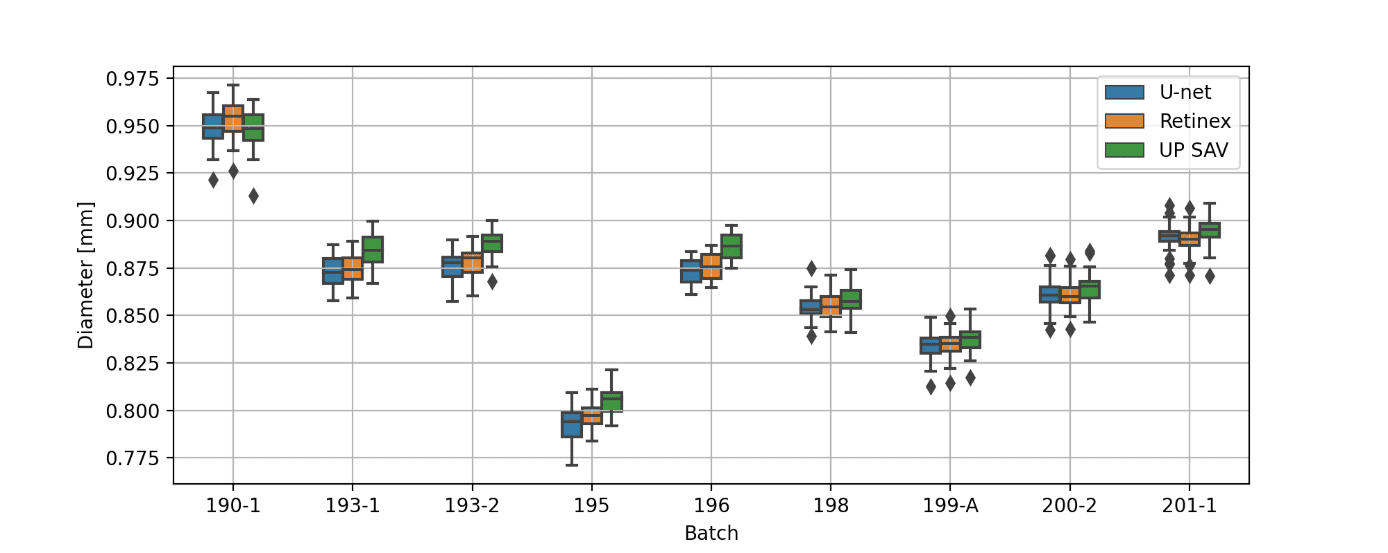
(a)
(b)
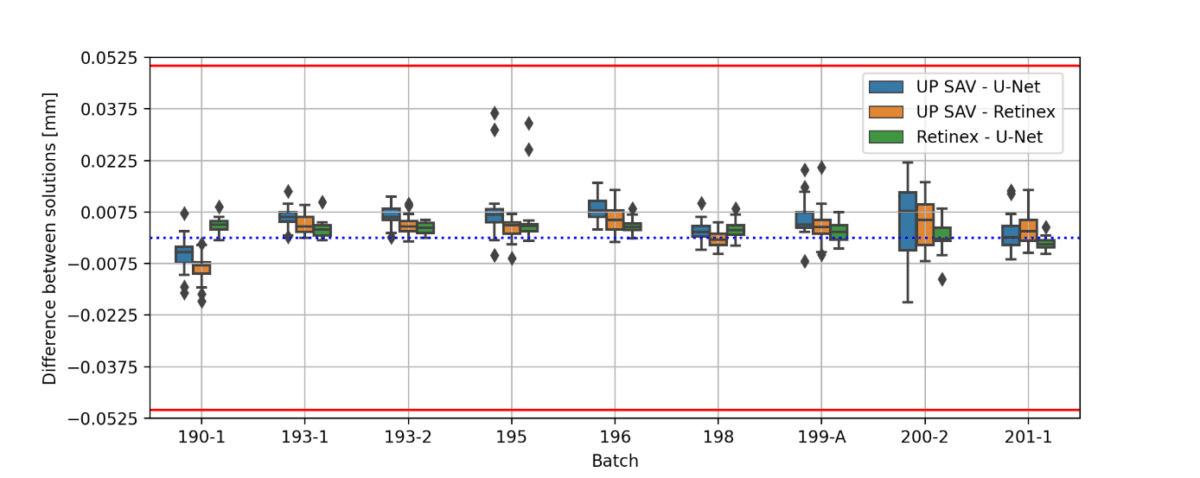
(c)
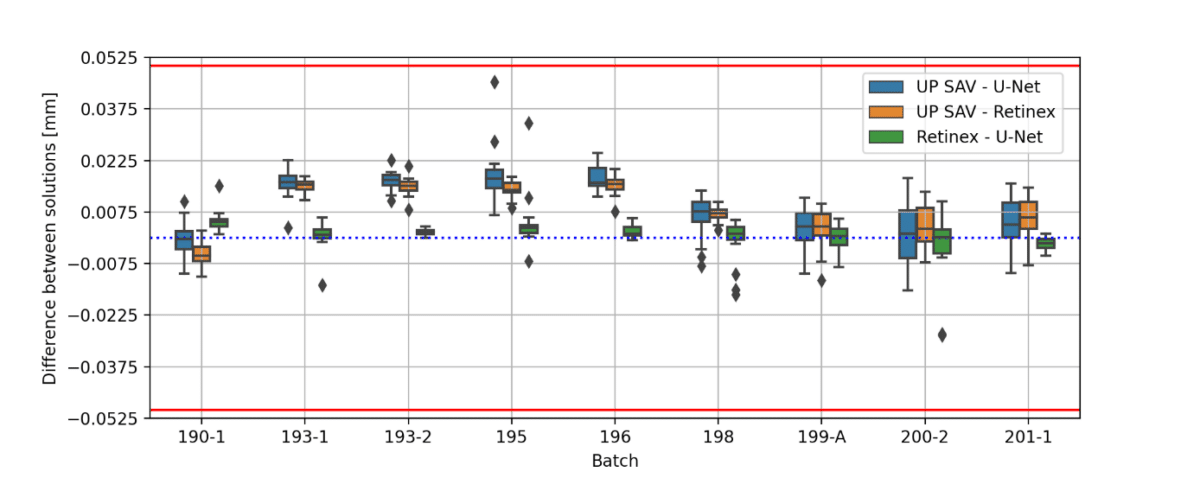
Figure 3: (a) microcapsule diameter for different batches (b) difference between the diameters of the fitted ellipse (first principal axis) and microcapsule (c) difference between the diameters of the fitted ellipse (second principal axis) and microcapsule. Red lines in (b) and (c) represents the threshold given by IP SAV. The images were obtained using 4.0 zoom.
All obtained results, except 4 images of batch 194 (ca 1.5%), are within the threshold defined by the IP SAV. As can be seen from Figure 3(a), the microcapsule diameters calculated using U-net and Retinex are in a good agreement to each other. The U-Net model performance can be significantly improved in future, either by the training set expansion or by additional post-processing. The agreement between the manual measurement and the U-Net/Retinex may be further improved by unifying the method of obtaining microcapsule structural parameters from binary masks.
The AI/ML model will be available as a cloud solution on the HPC systems of CSČ SAV. Additional investment into the HPC infrastructure of IP SAV will not be necessary. Production phase, which goes beyond the scope of the pilot solution, accounts for an integration of this approach into the desktop application.
References::
[1] https://github.com/ultralytics/yolov5
[2] https://www.kaggle.com/ultralytics/coco128
[3] https://github.com/heartexlabs/labelImg
[4] https://lmb.informatik.uni-freiburg.de/people/ronneber/u-net/
[5] https://docs.scipy.org/doc/scipy/reference/generated/scipy.ndimage.binary_fill_holes.html
[6] https://scikit-image.org/docs/stable/auto_examples/segmentation/plot_watershed.html
[7] https://scikit-image.org/docs/stable/api/skimage.measure.html
[8] D.J. Jobson, Z. Rahman, G.A. Woodell, IEEE Transactions on Image Processing 6 (7) 965-976, 1997.